May 19, 2016 – A team led by the Department of Energy’s Oak Ridge National Laboratory used neutron analysis to better understand a protein implicated in the replication of HIV, the retrovirus that causes AIDS. The enzyme, known as HIV-1 protease, is a key drug target for HIV and AIDS therapies.
Researchers from ORNL, Georgia State University and the Institut Laue-Langevin in France used neutron crystallography to uncover details of interactions of hydrogen bonds at the enzyme’s active site, revealing a pH-induced proton ‘hopping’ mechanism that guides its activity. The team discussed the findings in a paper published in the journal Angewandte Chemie.
Understanding the enzyme’s structure and function at the atomic level, including the location and movement of hydrogen atoms, is vital for understanding drug resistance and guiding rational drug design.
HIV-1 protease is responsible for the maturation of virus particles into infectious HIV virions, which ultimately leads to the development of AIDS. Without effective HIV-1 protease activity, HIV virions remain non-infectious, so the disruption of HIV-1 protease activity is a key target for successful antiretroviral therapy (ART) drugs that attack the virus itself.
The use of X-ray crystallography to study the structures of HIV-1 protease and drug complexes has led to the design of effective, commercially available ART drugs, but x-rays cannot determine the positions of mobile hydrogen atoms and protons. Neutron crystallography, however, can reveal these hydrogen-bonding interactions, which play a key role in how effectively a drug binds to its target.
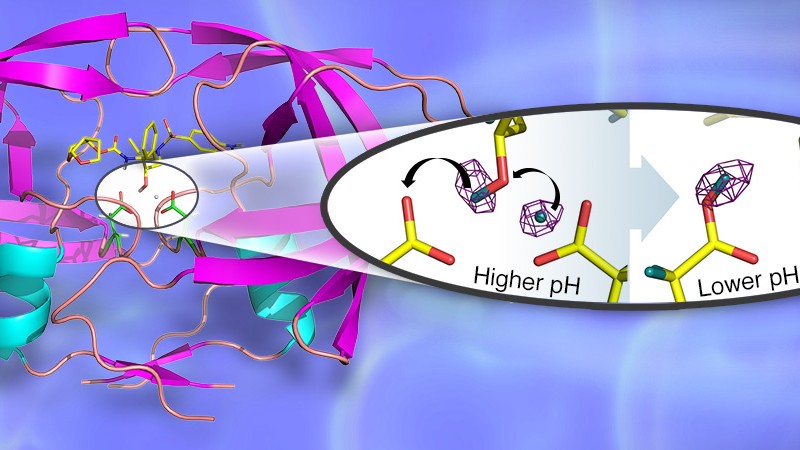
The ORNL-ILL-Georgia State research team used neutron crystallography to probe the structure of HIV-1 protease in complex with the clinical inhibitor Darunavir. The researchers combined neutron diffraction data from the IMAGINE instrument at ORNL’s High Flux Isotope Reactor (HFIR), a DOE Office of Science User Facility, and the LAD-III instrument at ILL, to uncover details of the hydrogen-bonding interactions in the active site and reveal ways to enhance drug binding and reduce drug resistance. The group also examined how the enzyme’s catalytic activity responds to changes in pH (acidity) levels.
By determining structures at different pHs, the group directly observed the positions of hydrogen atoms before and after a pH-induced two-proton transfer between the drug and enzyme. The proton transfer, triggered by electrostatic effects arising from proton uptake by surface residues from solution, resulted in the proton configuration that is critical for the catalytic activity.
“These results highlight that neutrons represent a superb probe to obtain structural details for proton transfer reactions in biological systems,” said ILL instrument scientist Matthew Blakely.
“Darunavir’s structure allows it to create more hydrogen bonds with the protease active site than most drugs of its type, while the backbone of HIV-1 protease maintains its spatial conformation in the presence of mutations,” said ORNL instrument scientist Andrey Kovalevsky. “This means Darunavir-protease interaction is less likely to be disrupted by a mutation. Given these characteristics, Darunavir is an excellent therapy target to refine and therefore enhance HIV treatment.”
To read the full story, click here.